Unearthing the Secrets of AREA_sample_R503: What One Microbial Genome Can Teach Us
When the term “genome” comes up, our thoughts often drift to humans or popular pets like dogs and cats. However, we seldom consider the rich tapestry of genetic life lurking just beneath the surface. In my recent project, I delved into the genome of a fascinating microorganism, known as AREA_sample_R503. This small but mighty microbe serves as a powerful lens into the hidden biodiversity surrounding us. As part of a larger research initiative, this genome contains a remarkable narrative intricately woven within its DNA.
Why This Genome Matters
AREA_sample_R503 was isolated from a soil sample taken in an agricultural setting, where the presence of microbial life is essential for plant health, nutrient cycling, and disease suppression. Although we haven’t assigned an official species name to this microbe yet, our genome sequencing efforts have enabled us to identify its closest relatives and make predictions about its role in the soil ecosystem.
Utilizing genome annotation tools such as PATRIC and RAST, we discovered that AREA_sample_R503 is part of the phylum Actinobacteria, a group renowned for its antibiotic production and ability to break down organic matter. Notably, this genome features a unique cluster of genes related to secondary metabolite production—these are compounds that can inhibit rival microbes or positively affect plant growth.
Big Ideas from a Small Genome
What’s truly fascinating about this project is how one genome can reflect the larger patterns of microbial life. AREA_sample_R503 had genes that hint at nitrogen fixation, which is the ability to convert nitrogen from the air into a form that plants can use—an essential function in sustainable agriculture. We also found several antibiotic resistance genes, which may indicate that this microbe is either defending itself from natural threats or influenced by human activities like overuse of antibiotics in farming.
This genome also revealed insights into carbon cycling, with enzymes that help break down complex plant materials. This positions AREA_sample_R503 as a potential key player in soil health, contributing to the breakdown of organic matter and the release of nutrients.
Findings
I was able to use different tools to analyze different aspects of my genome. We used CARD to find that one of the mechanisims is efflux and it encodes one beta-lactamase gene and possibly 4 classes of antibiotic resistance genes.
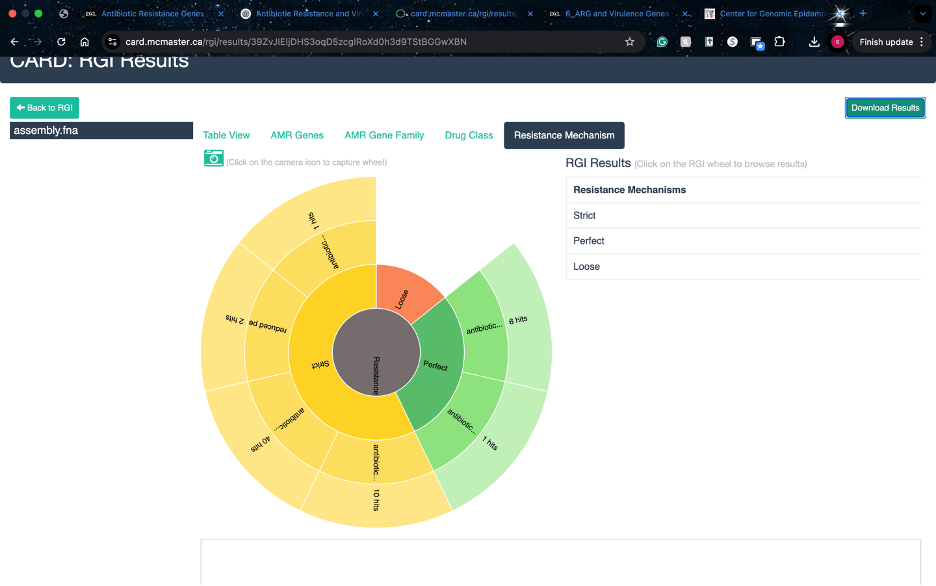
CARD Analysis
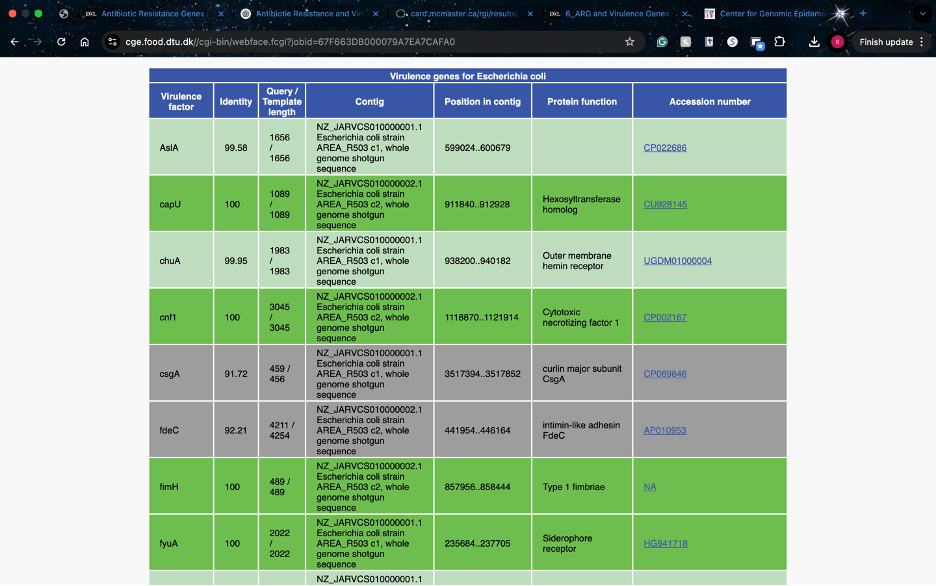
Virulence Finder
Why This Work Matters for the Future
Studying genomes such as AREA_sample_R503 allows scientists to gain deeper insights into the vital roles that microbes play in our ecosystems. These microscopic organisms contribute significantly to the health of our soil, the purity of our water, and even the regulation of our climate. An intriguing area for further research could be exploring whether this particular microbe is capable of producing novel antibiotics or compounds that promote plant growth, ultimately helping to lessen our dependence on chemical fertilizers and pesticides.
This research plays a key role in the worldwide mission to chart microbial diversity. Programs such as GTDB-Tk are empowering scientists to reclassify microorganisms through their evolutionary connections rather than solely their physical traits or growth techniques. This approach highlights the vast, unexplored territories of the microbial realm.
Final Thoughts
Collaborating with AREA_sample_R503 has demonstrated that a single microbe can provide profound insights into evolution, sustainability, and biotechnology. As we advance in decoding these genomes, we’re revealing opportunities to transform our approaches to food production, healthcare, and our understanding of life on Earth.Working with AREA_sample_R503 has shown me that even a single microbe can offer insights into evolution, sustainability, and biotechnology. As we continue to decode these genomes, we’re uncovering the potential to reshape how we grow food, treat disease, and understand life on Earth.
Whether it’s cleaning up pollutants or producing life-saving drugs, microbes like AREA_sample_R503 are silent heroes in our natural world—hidden in the soil, but vital to our future.
Reflection
My Signature Experience truly deepened my passion for health and sparked an enthusiasm for research. Throughout this project, I had the opportunity to work with a variety of bioinformatic tools that not only enhance my qualifications for future research and clinical roles but also boost my confidence. These tools helped me develop my writing, critical thinking, and analytical skills in a significant way.
I used to feel uncertain about stepping into the biomedical field, but completing several successful projects has truly solidified my sense of belonging. I demonstrated my attention to detail during the data analysis, particularly with mobile genetic elements, and I honed my ability to synthesize results from different tools into a cohesive conclusion. This skill is invaluable for my goal of becoming a researcher, as it trains us to gather all necessary information, cinstruct a concise and orderly exoeriment, and finally interperate and publish results.
Moreover, researchers must evaluate all results to arrive at a diagnosis that aligns with the overall clinical picture. Overall, this experience has equipped me for the complex challenges ahead, whether in a lab or hospital setting, and I’m eager to embrace those opportunities!